Abstract
Introduction: Acute lymphoblastic leukemia (ALL) is the most common pediatric cancer and the leading cause of cancer-related mortality in pediatric populations. Despite intensified treatment regimens, 15% of ALL patients relapse. Tumor heterogeneity has been implicated in many cancers as a prognostic feature for developing therapy resistance and relapse. The lack of successful therapies for the treatment of relapsed ALL makes the investigation of ALL progression fundamental. ALL is characterized by the accumulation of immature cells in the lymphoid lineage. This cell arrest is driven by oncogenic rearrangements that result in the aberrant expression of oncogenes. Differently from acute myeloid leukemia, which is originating from a leukemic stem cell, ALL is thought to lack hierarchical organization complicating targeted therapies directed toward persistent leukemia-initiating cells. Xenograft models of B-ALL suggest that subclones with differentiated and less-differentiated phenotypes engraft similarly and therefore can both be considered leukemia-initiating cells (LIC). However, for T-ALL this is less well studied and there are conflicting findings in the literature. Moreover, the relatively straightforward developmental trajectory of normal T-cell development is extensively studied, making T-ALL the perfect model to study ALL progression and heterogeneity. Therefore, we aim to improve our understanding of T-ALL tumor initiation and progression by studying phenotypic heterogeneity and inferring their ancestral hierarchy using single-cell DNA sequencing.
Methods: We characterized 29 diagnostic T-ALL samples (enriched for >90% blasts) for sub-clonal differentiation stages using 19 parameter flow cytometry. Additionally, we used Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-Seq) for 11 patients and single-cell whole genome sequencing (sc-WGS) for two patients to correlate the immunophenotypic subclones identified at diagnosis and infer the timing and ancestral hierarchy of T-ALL at diagnosis.
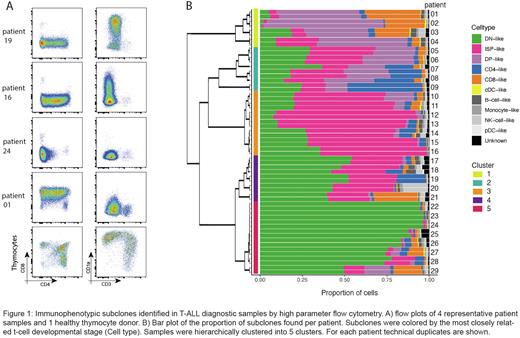
Results: We have identified phenotypic intra-tumor heterogeneity in 24/29 (83%) T-ALL diagnostic samples using high parameter flow cytometry (Fig. 1A). The sub-clonal phenotypes each reflect an arrest in different stages of healthy T-cell differentiation. Hierarchical clustering revealed 5 clusters of patients showing similar subclones and proportions of cells per subclone (Fig. 1B). Our CITE-seq analysis performed for 11 T-ALL patients included 204 antibodies and nicely reproduced these initial findings. Furthermore, we showed in an ETP-ALL sample, in which we did not find phenotypic heterogeneity based on 19 parameter flow cytometry, immunophenotypic subclones of CD34+CD62L+/CD34+CD62L- phenotype. The phenotypic heterogeneity was less pronounced in the transcriptomic data suggesting ALLs have a continuum of differentiation states among subclones. We are currently studying the ancestral hierarchy of phenotypic subclones we identified in T-ALL patients samples by sc-WGS analysis. By studying passenger mutations, we can construct phylogenetic trees that reflect the timing, hierarchy, and organization of leukemic cells.
Conclusion: One of the greatest challenges in treating ALL is reducing relapse. Our findings will provide novel insights into the hierarchy and sub-clonal progression of ALL which will help to identify ALL initiating cells that may re-initiate leukemia after therapy.
Disclosures
Meijerink:Acerta Pharma: Current Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal